T-Test
In short: T-tests can be used to investigate whether there is a significant difference between two groups regarding a mean value (two-sample t-test) or the mean in one group compared to a fixed value (one-sample t-test). In the following article, the concept and purpose of the t-test, assumptions, the implementation in R as well as multiple variants for different conditions will be covered.
Contents
General Information
Let us start by looking at the basic idea behind a two-tailed two-sample t-test. Conceptually speaking we have two hypothesis:
H0: Means between the two samples do not differ significantly. H1: Means between the two samples do differ significantly.
In mathematical terms (μ1 and μ2 denote the mean values of the two samples):
H0: μ1 = μ2
H1: μ1 ≠ μ2.
Here an example to illustrate this. The farmers Kurt and Olaf grow sunflowers and wonder who has the bigger ones. So they each measure a total of 100 flowers and put the values into a data frame.
# Create a dataset set.seed(320) kurt <- rnorm(100, 60.5, 22) olaf <- rnorm(100, 63, 23)
Our task is now to find out whether means values differ significantly between two groups.
# perform t-test t.test(kurt, olaf) ## Output: ## ## Welch Two Sample t-test ## ## data: kurt and olaf ## t = -1.5308, df = 192.27, p-value = 0.1275 ## alternative hypothesis: true difference in means is not equal to 0 ## 95 percent confidence interval: ## -11.97670 1.50973 ## sample estimates: ## mean of x mean of y ## 57.77072 63.00421
Cool. We performed a t-test and got a result. But how can we interpret the output? The criterion to consult is the p-value. This value represents the probability of the data given that H0 is actually true. Hence, a low p-value indicates that the data is very unlikely if H0 applies. Therefore, one might reject this hypothesis (in favor of the alternative hypothesis H1) if the p-value turns out to be below a certain threshold (α , usually set prior testing), which is often set to 0.05 and usually not larger than 0.1. In this case, the p-value is greater than 0.1. Therefore, the probability of H0 is considered to be “too large” to reject this hypothesis. Hence, we conclude that means do not differ significantly, even though we can say that descriptively the sample mean of Olaf’s flowers is higher.
There are multiple options to fine-tune the t-test if one already has a concrete hypothesis in mind concerning the direction and/or magnitude of the difference. In the first case, one might apply a one-tailed t-test. The hypotheses pairs would change accordingly to either of these:
H0: μ1 ≥ μ2
H1: μ1 < μ2
or
H0: μ1 ≤ μ2
H1: μ1 > μ2
Note that the hypotheses need to be mutually exclusive and H0 always contains some form of equality sign. In R, one-tailed testing is possible by setting alternative = "greater" or alternative = "less". Maybe Olaf is the more experienced farmer so we have already have à priori the hypothesis that his flowers are on average larger. This would refer to our alternative hypothesis. The code would change only slightly:
t.test(kurt, olaf, alternative = "less") ## Output: ## ## Welch Two Sample t-test ## ## data: kurt and olaf ## t = -1.5308, df = 192.27, p-value = 0.06373 ## alternative hypothesis: true difference in means is less than 0 ## 95 percent confidence interval: ## -Inf 0.4172054 ## sample estimates: ## mean of x mean of y ## 57.77072 63.00421
As one can see, the p-value gets smaller, but it is still not below the “magic threshold” of 0.05. The question of how to interpret this result might be answered differently depending on whom you ask. Some people would consider the result “marginally significant” or would say that there is “a trend towards significance” while others would just label it as being non-significant. In our case, let us set α
= 0.05 for the following examples and call a result “significant” only if the p-value is below that threshold.
It is also possible to set a δ indicating how much the groups are assumed to differ.
H0: μ1 - μ2 = δ
H1: μ1 - μ2 ≠ δ
H0: μ1 - μ2 ≥ δ
H1: μ1 - μ2 < δ
H0: μ1 - μ2 ≤ δ
H1: μ1 - μ2 > δ
To specify a δ, set mu = *delta of your choice*. If one sets a mu and only specifies one group, a one-sample t-test will be performed in which the group mean will be compared to the mu.
Assumptions
Like many statistical tests, the t-test builds on a number of assumptions that should be considered before applying it.
- The data should be continuous or on an ordinale scale.
- Normally distributed data. However (according to the central limit theorem), the empirical distribution function of the standardised means converges towards the standard normal distribution with growing sample size. Hence, this prerequisite can be assumed to be met with a sufficiently large sample size (≥ 30).
- Data is drawn randomly from a representative sample.
- For Student’s t-test, equal variances in the two groups are required. However, by default, the built in function t.test() in R assumes that variances differ (Welch t-test). If it is known that variances are equal, one can set var.equal = TRUE, which will lead to a Student’s t-test being performed instead.
Paired t-test
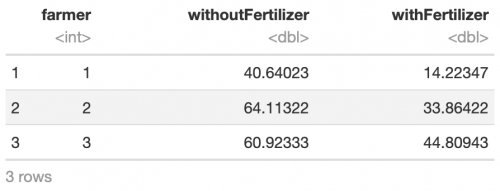
In many cases one has to do with groups that are not independent of each other. This is the case for example in within-subject designs where one compares data between the time points but within one participant. In this case, a paired t-test should be the means of choice. In R, this option can be chosen by simply setting paired = TRUE. It has the advantage of increasing statistical power, that is to say the probability of finding an effect (getting a significant result) if there actually is one. Let’s modify our example: One hundred farmers try out a new fertiliser. Now we want to investigate whether the height of flowers treated with this fertiliser differs significantly from naturally grown ones. In this case, in each row of the dataset there is the mean value of flower size per farmer for each treatment:
# Create dataset
set.seed(320)
farmer <- c(1:100)
withFertilizer <- rnorm(100, 60, 22)
withoutFertilizer <- rnorm(100, 64, 17)
ds <- data.frame(farmer,
withoutFertilizer,
withFertilizer)
# inspect the data
head(ds, 3)
# perform t-test t.test(ds$withoutFertilizer, ds$withFertilizer, paired = T) ## Output: ## ## Paired t-test ## ## data: ds$withoutFertilizer and ds$withFertilizer ## t = 2.2801, df = 99, p-value = 0.02474 ## alternative hypothesis: true difference in means is not equal to 0 ## 95 percent confidence interval: ## 0.8737431 12.5910348 ## sample estimates: ## mean of the differences ## 6.732389
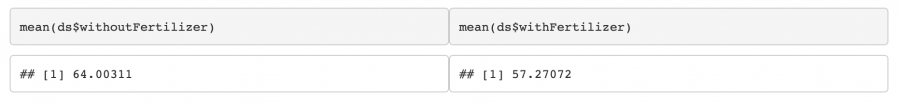
In this case, the p-value is below our chosen α = 0.05. Therefore, we can reject H0 and conclude that there is probably a significant difference between means. But be careful! One might suspect that the fertiliser leads to an increased growth, but by looking at the mean values we realize that here, the contrary seems to be the case:
Multiple testing
When performing a large number of t-tests (e.g. when comparing multiple groups in post-hoc t-tests after performing an ANOVA), one should keep in mind the issue of alpha-error-cumulation resulting in the “problem of multiple comparison”. It states that with increasing number of tests performed the probability of making a Type-I error (falsely rejecting the null-hypothesis in favor of the alternative hypothesis) increases. To account for this risk, p-values should be adjusted. For instance, one might use the function pairwise.t.test(x = *response vector*, g = *grouping vector or factor*). It performs t-tests between all combinations of groups and by default returns bonferroni corrected p-values. The adjustment method can be chosen by setting p.adjust.method equal to the name of a method of choice. For options see ?p.adjust. An alternative would be to adjust p-values “manually”:
p.adjust(p = result_t.test[3], # p = object in which result of t.test is stored
method = "bonferroni", # method = p-adjustment method
n = 3) # n = number of tests
An additional tip for R Markdown users
If you are using R Markdown you may use inline code to automatically print nicely formatted results (according to apa) of a t-test including degrees of freedom, test-statistic, p-value and Cohen’s d as measure for effect size. You would need to use the funcion t_test (similar to t.test) and store the result in an object…
result <- t_test(ds$withoutFertilizer,
ds$withFertilizer,
paired = T)
# the inline-code would look like this:
# `r apa(result)`
…that can then be called inline (see above). The result will look like this: t(99) = 2.28, p = .025, d = 0.23. Note that the functions t_test() and apa() require the package “apa”.